For loops are very convenient. With a combination of a for loop and some if-else statements you can do just about anything. There’s just one problem though, for loops are kinda slow. There are many ways to speed up the code, the best usually, in the data science world, to use vectorized NumPy or Pandas solutions. Yet if you are stuck with a for loop, you are in what’s called an ’embarrassingly parallel’ situation. That is, a for loop can easily be performed by many workers, not just one, and thus be made much faster.
First off, some background. There are two related forms of parallelization: multithreading and multiprocessing. Multithreading has lower overhead and uses shared memory, while multiprocessing uses different environments entailing more overhead both in compute time and memory. For reasons of simplicity and robustness, Python was built with the Global Interpreter Lock (GLI). It prevents multithreading, but allows multiprocessing. Except Python does allow multithreading, as long as your code doesn’t actually use any Python code (hint: Cython and stuff). Confused yet? The conclusion of all this is that in Python, unless you are a highly advanced user, you can use multiprocessing (but not threading) to help make your code faster.
Things get even more confusing because your code may already be using multiprocessing or multithreading. Here’s where you might expect to see it:
- BLAS level multithreading, ie in Intel MKL
- Lower-level library multithreading/multiprocessing ie in scikit-learn
- Your program multiprocessing
The BLAS (Basic Linear Algebra Subprogram) varies depending on your system, and some of them have automatic multithreading. The BLAS takes care of feeding linear algebra instructions to the CPU and is what actually performs your matrix calculations regardless of whether you are in Python, Matlab, or Excel. Intel MKL (Intel’s optimized BLAS) is built into Anaconda distributions of Python and will use multithreading by default. You can check your numpy’s BLAS in np.show_config(). By the way, if you are not on an Intel system, avoid Intel MKL as it is designed to throttle itself if it fails to detect an Intel CPU. The next level up, packages like scikit-learn often have the option to use parallelization. Some packages have multiprocessing on by default.
It’s important to run you code in an un-parallelized way first and see if CPU use is above what you would expect for a single core. Say you are on a four core system, and some sections of your code already use 4 threads. If you in turn add parallelization, your program may well be trying to ask for 4 * 4 cores/threads. Going above your expected core count can lead to the code actually getting slower as processes fight among themselves for resources. It may also outright crash your computer. Crashing in weird and funky ways is all too often what happens when CPU or GPU parallelization is attempted.
At this point, you are still using a for loop and you have checked to see that your current for loop uses just around 1 core of CPU for itself. To make it parallel, you have three basic choices: concurrent.futures and the multiprocessing libraries built into Python 3, and joblib – a relatively minimal third-party package used by (indeed built for) scikit-learn.
The most flexible and most well-documented option is multiprocessing. The newer built-in option is concurrent.futures, and it is a little faster than multiprocessing. On the other hand, joblib is the smartest option – automatically reusing threads, attempting to reasonably compensate for BLAS level hyperthreading, and also allowing integration with Dask for distribution across a cluster.
Multiprocessing has its problems. It is a bit picky about how it runs on Windows, and it doesn’t work consistently in interactive (iPython) sessions. One thing to very much be aware of is that is takes a while to recruit a worker pool. On the two systems I have been testing on here, I saw it vary between 1s and 10s delays (with no clear correlation with why sometimes it was faster or slower). That means your for loop needs to run > ~20 seconds long before you will notice any return. Indeed, I would not usually trouble with multiprocessing unless your task is running for minutes, hours, or better yet days.
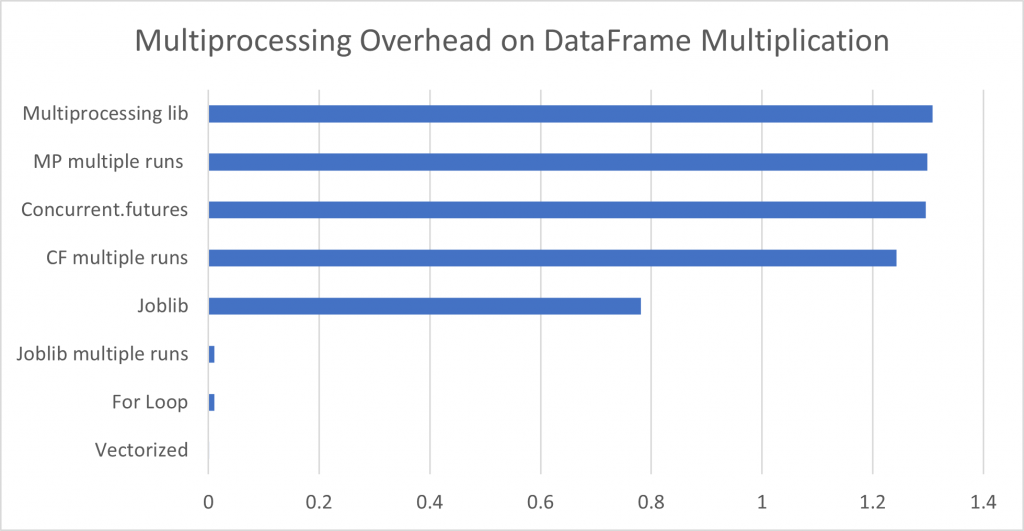
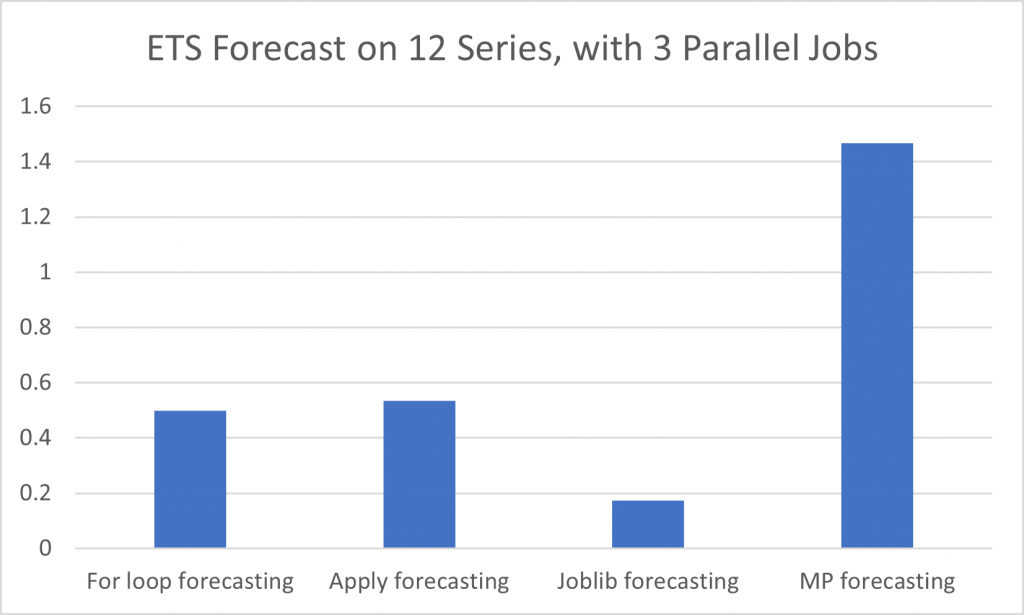
Running a comparison of DataFrame multiplication using a DataFrame of 12 columns, each with rows of the identical 1 to 2000 sequence, it is quite obvious that a vectorized solution is three orders of magnitude faster than multiprocessing because the overhead for creating the pool is so great. Joblib multiprocessing was nearly identical to a regular for loop as it benefits from automatically reusing an existing process pool. Yet on a much more computationally intensive problem, that of an ETS forecast on the same dataset, multiprocessing begins to stand out, as a 3 process job is almost exactly 3x faster than a conventional for loop (in this case joblib was running on an existing pool, hence minimal overhead).
Side-note on hyperthreading: many CPUs use ‘hyperthreading’ which basically doubles the thread count so there are 2 threads for every 1 core. This allows the CPU overall to be faster, but it also confuses many programs into thinking you have twice as many CPUs as actually you have. For example, many systems have 4 cores but os.cpu_count() will report 8 cores – confused by the number of threads provided by hyperthreading.
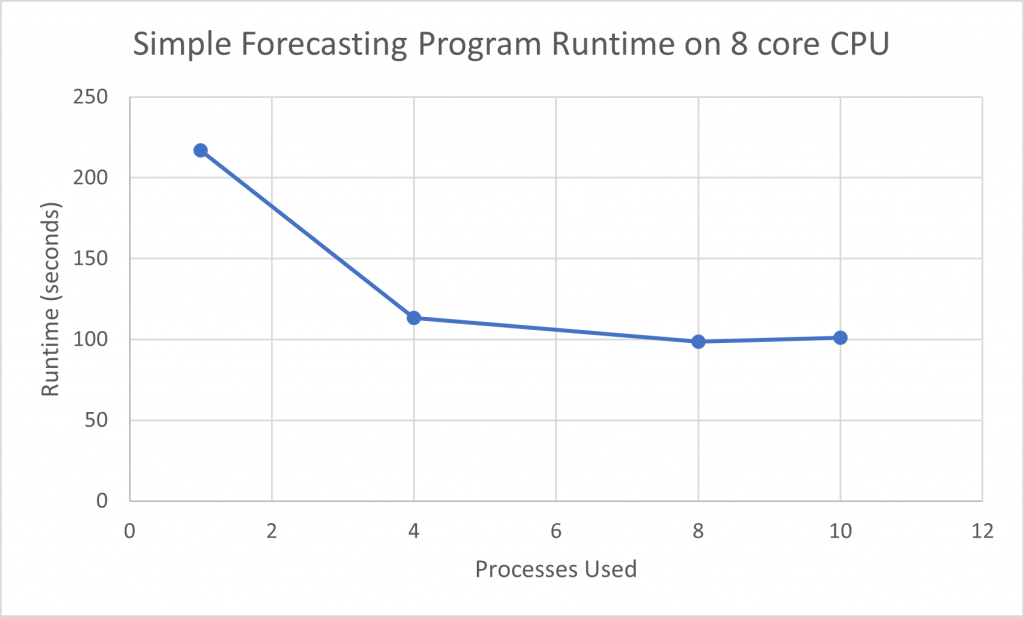
In another example with 9 time series and running a number of models, there is little performance difference between using 4 processes and using 8 processes. While using 4 processes cut more than half of the runtime off a single CPU run, the remainder of the code in this program, running sequentially, will not see any performance gain, hence the ‘leveling out’ observed.
Overall, my recommendation, if multiprocessing is appropriate, is to use joblib for the best performance utilizing multiple parallelized loops, but for a single multiprocessing loop, either of the built in packages multiprocessing or concurrent.futures is more convenient, not requiring an external dependency.
Further Readings:
https://homes.cs.washington.edu/~jmschr/lectures/Parallel_Processing_in_Python.html
https://conference.scipy.org/proceedings/scipy2018/anton_malakhov.html
Code for Examples
mpsource.py
import numpy as np
import pandas as pd
from statsmodels.tsa.holtwinters import ExponentialSmoothing
def series_by_column(lst, col):
out = (col + 2) ** 2
lst.append(out)
def series_by_column_cf(col):
return (col + 2) ** 2
def df_by_column(df, col):
out = (df[col] + 2) ** 2
return out
def forecast_by_column(df, col):
current_series = df[col]
series_name = current_series.name
esModel = ExponentialSmoothing(
current_series,
damped=False,
trend="additive",
seasonal=None,
seasonal_periods=7,
freq="D",
).fit()
srt = current_series.shape[0]
esPred = esModel.predict(start=srt, end=srt + 10)
esPred = pd.Series(esPred)
esPred.name = series_name
return esPred
def forecast_by_series(lst, col):
current_series = col
series_name = current_series.name
esModel = ExponentialSmoothing(
current_series,
damped=False,
trend="additive",
seasonal=None,
seasonal_periods=7,
freq="D",
).fit()
srt = current_series.shape[0]
esPred = esModel.predict(start=srt, end=srt + 10)
esPred = pd.Series(esPred)
esPred.name = series_name
lst.append(esPred)
def forecast_from_series(col):
current_series = col
series_name = current_series.name
esModel = ExponentialSmoothing(
current_series,
damped=False,
trend="additive",
seasonal=None,
seasonal_periods=7,
freq="D",
).fit()
srt = current_series.shape[0]
esPred = esModel.predict(start=srt, end=srt + 10)
esPred = pd.Series(esPred)
esPred.name = series_name
return esPredmptest.py
import numpy as np
import pandas as pd
import timeit
from mpsource import (
series_by_column,
series_by_column_cf,
df_by_column,
forecast_by_column,
forecast_by_series,
forecast_from_series
)
n_jobs = 3
len_idx = 2000
ltrs = ["A", "B", "C", "D", "E", "F", "G", "H", "I", "J", "K", "L"]
idx = pd.date_range(start="2020-01-01", periods=len_idx, freq="D")
df = pd.DataFrame(
np.linspace(
np.repeat([1], len(ltrs)), np.repeat([len_idx], len(ltrs)), num=len_idx, axis=0
),
columns=ltrs,
index=idx,
)
df = df.astype(int)
col_order = df.columns
# For loop no parallel
start_time_for = timeit.default_timer()
out_list = []
for col in df.columns:
out_list.append((df[col] + 2) ** 2)
full_df = pd.concat(out_list, axis=1)
elapsed_for = timeit.default_timer() - start_time_for
# This is how you'd normally do a simple thing like this.
start_time_pd = timeit.default_timer()
full_df = (df + 2) ** 2
elapsed_pd = timeit.default_timer() - start_time_pd
start_time_cf = timeit.default_timer()
series_list = [df[col] for col in df.columns]
if __name__ == "__main__":
from concurrent.futures import ProcessPoolExecutor
from functools import partial
with ProcessPoolExecutor(max_workers=n_jobs) as executor:
res = executor.map(series_by_column_cf, series_list)
dfs = pd.concat(res, axis=1)
dfs = dfs[col_order]
else:
print("not __main__!")
elapsed_cf = timeit.default_timer() - start_time_cf
start_time_mp = timeit.default_timer()
series_list = [df[col] for col in df.columns]
if __name__ == "__main__":
from multiprocessing import Pool, Manager
from functools import partial
with Pool(processes=n_jobs) as pool, Manager() as manager:
dfs_list = manager.list()
res = pool.map_async(partial(series_by_column, dfs_list), series_list)
res.wait()
dfs = pd.concat(dfs_list, axis=1)
dfs = dfs[col_order]
else:
print("not __main__!")
elapsed_mp = timeit.default_timer() - start_time_mp
# JOBLIB Parallelization
start_time_jl = timeit.default_timer()
if __name__ == "__main__":
cols = df.columns.tolist()
from joblib import Parallel, delayed
df_list = Parallel(n_jobs=n_jobs)(
delayed(df_by_column, check_pickle=False)(df, col) for (col) in cols
)
dfs_joblib = pd.concat(df_list, axis=1)
else:
print("not __main__!")
elapsed_jl = timeit.default_timer() - start_time_jl
runs = 3
# JOBLIB reuse of pool by default
start_time_jl2 = timeit.default_timer()
for i in range(runs):
if __name__ == "__main__":
cols = df.columns.tolist()
from joblib import Parallel, delayed
df_list = Parallel(n_jobs=n_jobs)(
delayed(df_by_column, check_pickle=False)(df=df, col=col) for (col) in cols
)
dfs_joblib = pd.concat(df_list, axis=1)
elapsed_jl2 = (timeit.default_timer() - start_time_jl2) / runs
# MP without any reuse of pool
start_time_mp2 = timeit.default_timer()
series_list = [df[col] for col in df.columns]
for i in range(runs):
if __name__ == "__main__":
from multiprocessing import Pool, Manager
from functools import partial
with Pool(processes=n_jobs) as pool, Manager() as manager:
dfs_list = manager.list()
res = pool.map_async(partial(series_by_column, dfs_list), series_list)
res.wait()
dfs = pd.concat(dfs_list, axis=1)
dfs = dfs[col_order]
elapsed_mp2 = (timeit.default_timer() - start_time_mp2) / runs
# CF without any reuse of pool
start_time_cf2 = timeit.default_timer()
series_list = [df[col] for col in df.columns]
for i in range(runs):
if __name__ == "__main__":
from concurrent.futures import ProcessPoolExecutor
from functools import partial
executor = ProcessPoolExecutor(max_workers=n_jobs)
res = executor.map(series_by_column_cf, series_list)
dfs = pd.concat(res, axis=1)
dfs = dfs[col_order]
elapsed_cf2 = (timeit.default_timer() - start_time_cf2) / runs
print(
f"Vectorized is {elapsed_pd}\nFor loop is {elapsed_for}\n\nMultiprocessing lib is {elapsed_mp}\nConcurrent.futures is {elapsed_cf}\nJoblib is {elapsed_jl}\n\nMP multiple runs {elapsed_mp2}\nCF multiple runs {elapsed_cf2}\nJoblib multiple runs {elapsed_jl2}\n "
)
# yes a linear reg would be fine on this data, but ETS needs more parallel
# For Loop Forecasting
start_time_for_f = timeit.default_timer()
out_list = []
for col in df.columns:
out_list.append(forecast_by_series(out_list, df[col]))
forecast_df = pd.concat(out_list, axis=1)
elapsed_for_f = timeit.default_timer() - start_time_for_f
# Joblib Forecasting
# additional arguments passed through joblib need to have explicity keywords=keyword_value
start_time_jl_f = timeit.default_timer()
if __name__ == "__main__":
cols = df.columns.tolist()
from joblib import Parallel, delayed
df_list = Parallel(n_jobs=n_jobs)(
delayed(forecast_by_column, check_pickle=False)(df=df, col=col) for (col) in cols
)
forecast_df_joblib = pd.concat(df_list, axis=1)
elapsed_jl_f = timeit.default_timer() - start_time_jl_f
# MP Forecasting
start_time_mp_f = timeit.default_timer()
series_list = [df[col] for col in df.columns]
if __name__ == "__main__":
from multiprocessing import Pool, Manager
from functools import partial
with Pool(processes=n_jobs) as pool, Manager() as manager:
dfs_list = manager.list()
res = pool.map_async(partial(forecast_by_series, dfs_list), series_list)
res.wait()
dfs = pd.concat(dfs_list, axis=1)
dfs = dfs[col_order]
elapsed_mp_f = timeit.default_timer() - start_time_mp_f
# Apply Forecast
start_time_apply_f = timeit.default_timer()
forecast_apply = df.apply(forecast_from_series, raw=False, axis=0)
elapsed_apply_f = timeit.default_timer() - start_time_apply_f
# give it time to get the rest of the printing out
import time
time.sleep(5)
print(
f"Vectorized is {elapsed_pd}\nFor loop is {elapsed_for}\n\nMultiprocessing lib is {elapsed_mp}\nConcurrent.futures is {elapsed_cf}\nJoblib is {elapsed_jl}\n\nMP multiple runs {elapsed_mp2}\nCF multiple runs {elapsed_cf2}\nJoblib multiple runs {elapsed_jl2}\n "
)
print(
f"\nFor loop forecasting {elapsed_for_f}\nApply forecasting {elapsed_apply_f}\nJoblib forecasting {elapsed_jl_f}\nMP forecasting {elapsed_mp_f}"
)
Pingback: Setting up and Optimizing Python for Data Science on Intel, AMD, and ARM (including Apple) Computers – Syllepsis